Cell-Based Functional Assays
We perform different cell-based functional assays and use different readouts to determine the effect of treatment with compounds on cells. Compounds can be small molecules, antibodies, peptides, liposomal vesicles, or small inhibitory RNAs.
Cell Viability Assays
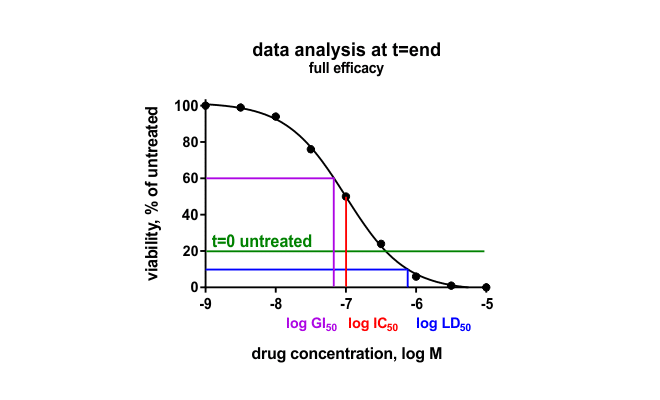
In Oncolines® profiling, the effect of compounds on cell viability is determined by measuring the intracellular content of ATP using ATPLite™ (PerkinElmer). Intracellular ATP is an indirect measure of cell number. The effect of compounds determined in our high-throughput cell viability assay correspond well with the effects determined in other cell-based assays, such as colony-forming unit assays but can also differ. Our standard Oncolines® profiling is performed in 2D. We also offer viability assays with cell cultures grown in 3D spheroids, and we offer alternative assay readouts for our proliferation assays.
ELISA or Multiplexing Immunoassay
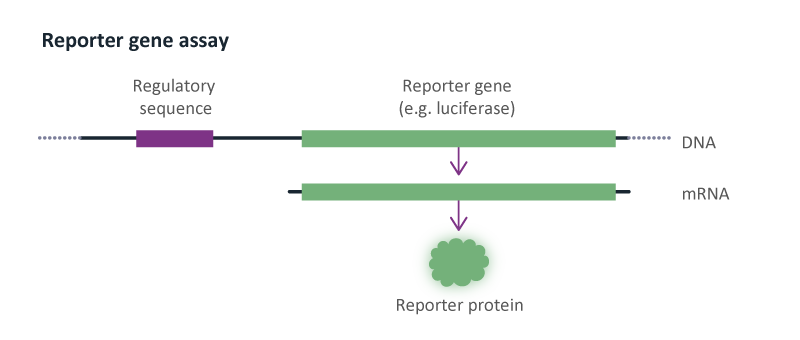
Reporter Gene Assays
Reporter gene assays are used to determine the effect of compounds on gene transcription. We have experience with luciferase and β-lactamase reporter genes. Reporter genes are used to study the effect of compounds on G protein-coupled receptors (GPCR) and nuclear hormone receptors.
Signal Transduction
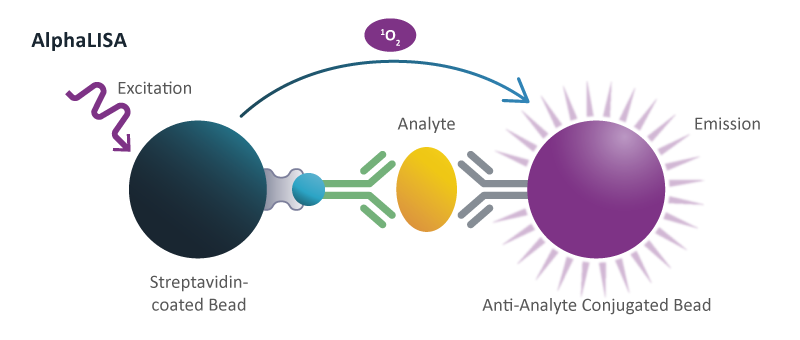
Different assays are used to study signal transduction, including high-throughput cAMP assays to study signaling downstream from G protein-coupled receptors, or phosphorylation assays to study the activity of kinases and phosphatases in cells. Phosphorylation of specific proteins can be measured in microtiter plate format assays using AlphaLISA®, a chemiluminescent proximity immunoassay. Effect of compounds on signal transduction, in particular phosphorylation of proteins, can also be studied by immunoblotting (Western).
Apoptosis
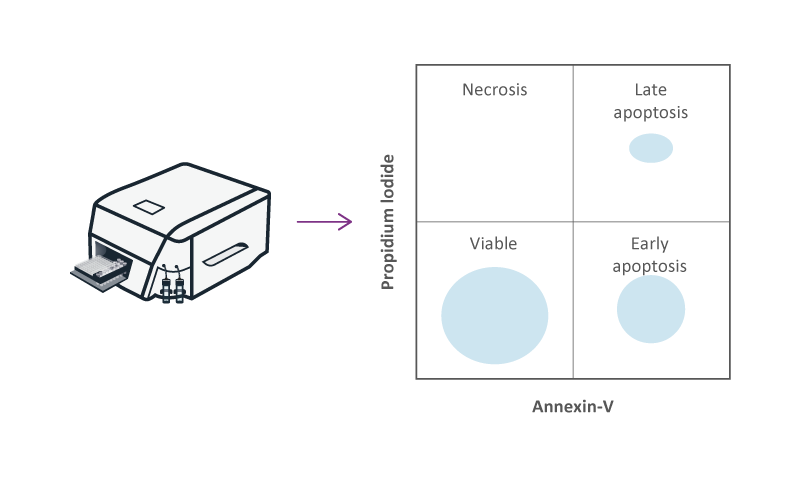
Apoptosis or “programmed cell death” is an orchestrated form of cell death, which is regulated by serine proteases. Apoptosis can be determined by measuring the activity of caspases in cells, or by the expression of Annexin V at the surface of cells. Measurement of Caspase 3 and 7 with the Caspase-Glo® assay is used to determine the effect of compounds on apoptosis in a microtiter plate-based assay (384-well assay). Flow cytometry is used to determine the expression of Annexin V at the surface of apoptotic cells using fluorescent-labeled antibodies. The effect of compounds on necrosis is measured using a fluorescent probe, such as propidium iodide (PI), that only binds to the DNA of cells with a leaky membrane.
Colony-Forming Unit Assays
In colony-forming unit (CFU) assays the effect of compounds on the ability of cells to form colonies is determined. Compared to cell viability assays, cells are seeded at lower density and treatment time with compound occurs over a longer period. If the effect of a compound is dependent on the number of cell cycles, its effect may be stronger in CFU assays in comparison to cell viability assays. CFU assays are also thought to be more representative of a condition of metastasis, which is initiated by the colonization of a tissue by one cancer cell that has spread from its original location in the body.
Co-Culture of Cancer and Immune Cells
Co-cultures are used to determine the effect of cancer cells on immune cells and are relevant in vitro assays to study cancer immunotherapy drugs. We have experience with two different types of co-culture assays. First, are immune cell-based assays, where immune cells are mixed with cancer cells in high-throughput cell viability assays. Specific T cell populations or other immune cells can be isolated by affinity chromatography and their effect studied in the presence of bispecific antibodies. Second, the effect of compounds on the interaction of immune cells with cancer cells can be studied by specific labeling of immune cells with fluorescent dye and flow cytometry. Immune cell proliferation is determined by measuring a fluorescent peak shift. Specific immune cell populations in the sample, such as T cells, are identified by labeling with specific antibodies against surface receptors.
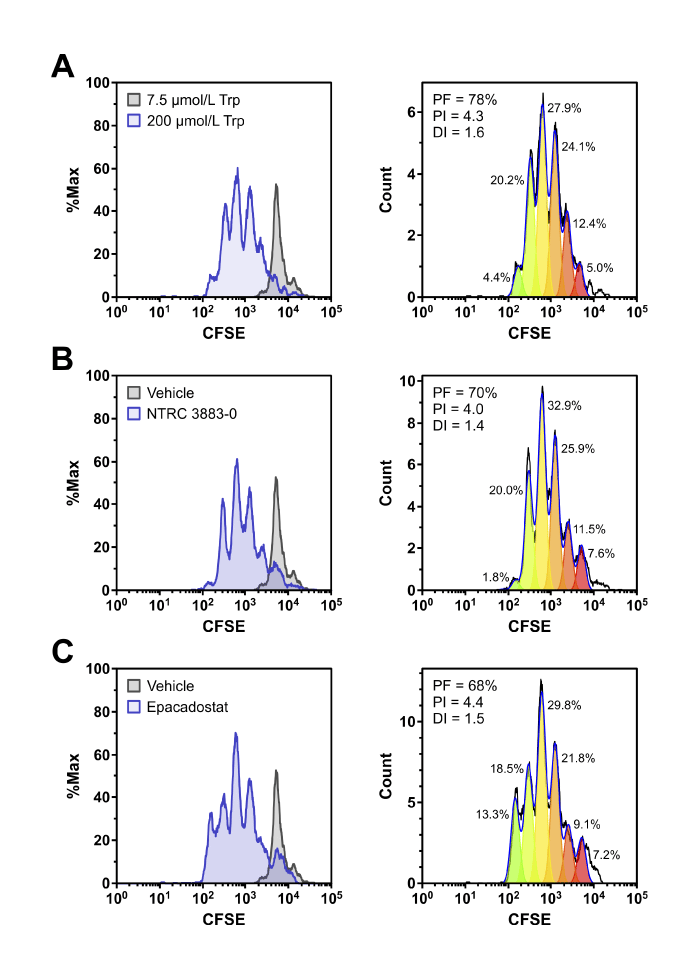
Co-culture assays of HEK-hIDO1 cells with lymphocytes from a healthy donor. Proliferation of CFSE-labeled CD8-positive T cells co-cultured for 5 days with HEK-hIDO1 cells upon addition of (A) 200 µM Trp as a positive control for T cell proliferation, (B) 10 µM NTRC 3883-0, and (C) 1 µM epacadostat. Each panel shows an overlay with CD8-positive T cells co-cultured for 5 days with HEK-hIDO1 cells upon addition of 7.5 µM Trp and vehicle (left), and the immune cell proliferation analysis (right). PF, precursor frequency (the percentage of cells from the original population that have proliferated); PI, proliferation index (the average number of cell divisions undergone by the proliferating cell population); and DI, division index (the average number of cell divisions undergone by the entire cell population). The percentages indicate the percentage of the total amount of cells in each generation.
Metabolite Analysis
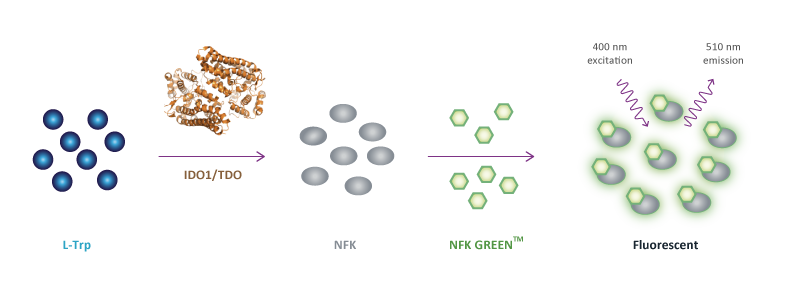
We have experience with the identification of (metabolites of) amino acids using liquid chromatography mass spectrometry. The levels of substrates and metabolites in cell culture supernatants, or samples of blood or ascites are quantitatively measured using calibration curves. For the tryptophan metabolizing enzymes indoleamine 2,3-dioxygenase 1 (IDO1) and tryptophan-2,3-dioxygenase (TDO), we have developed high-throughput fluorescent assays that can be used in 96-well, 384-well and 1536-well plates (NFK Green™ assay).
Oncolines Profiling with Immune Cells
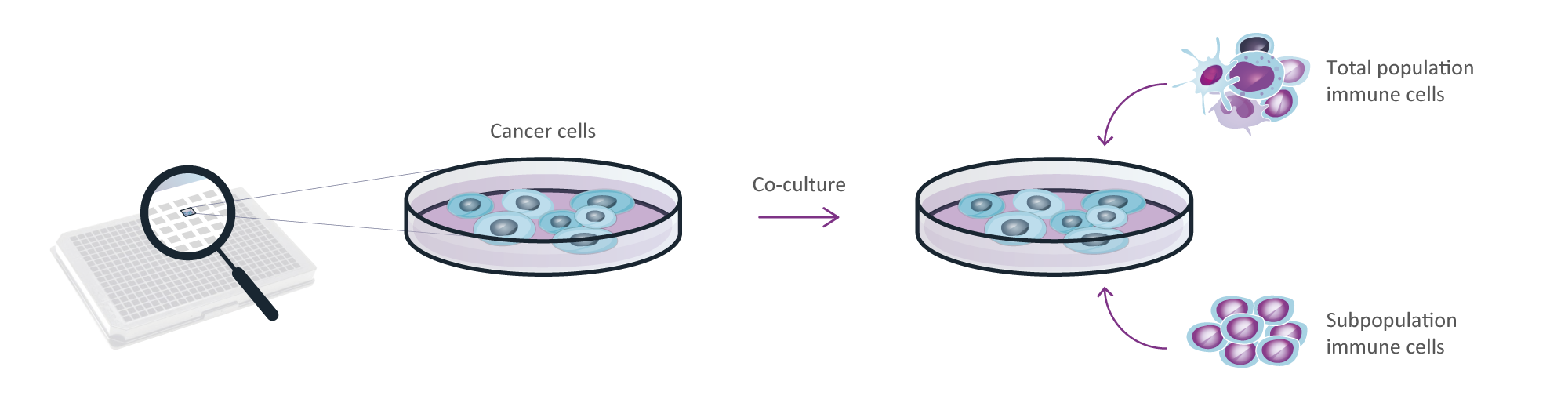
We offer a high-throughput Oncolines® Profiling panel with 102 cancer cell lines to study the effect of a single compound, or a combination of compounds, on cell proliferation. Additionally, immune cells (total population or a specific subpopulation) may be added to the Oncolines® Profiling panel to investigate the effect of co-cultures with these immune cells on proliferation of cancer cells in the presence of compound(s). With these experiments, we are also able to determine the production and secretion of cytokines after contact between the cells compared to mono-cultures by an ELISA or multiplex immunoassay. Moreover, the effect of engineered immune cells (e.g., T cells) can be tested on a broad panel of cancer cell lines.