Quantitative Polymerase Chain Reaction (qPCR)
Principle
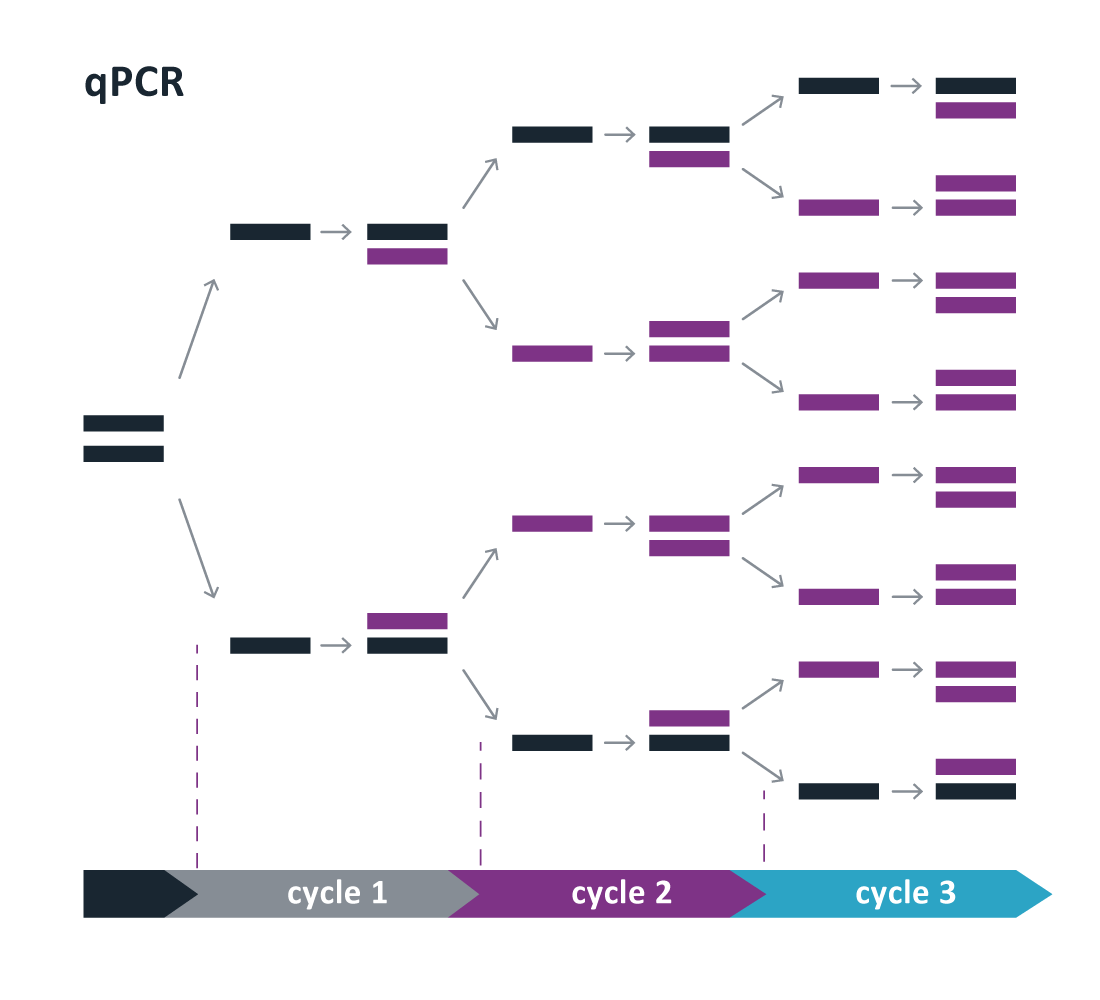
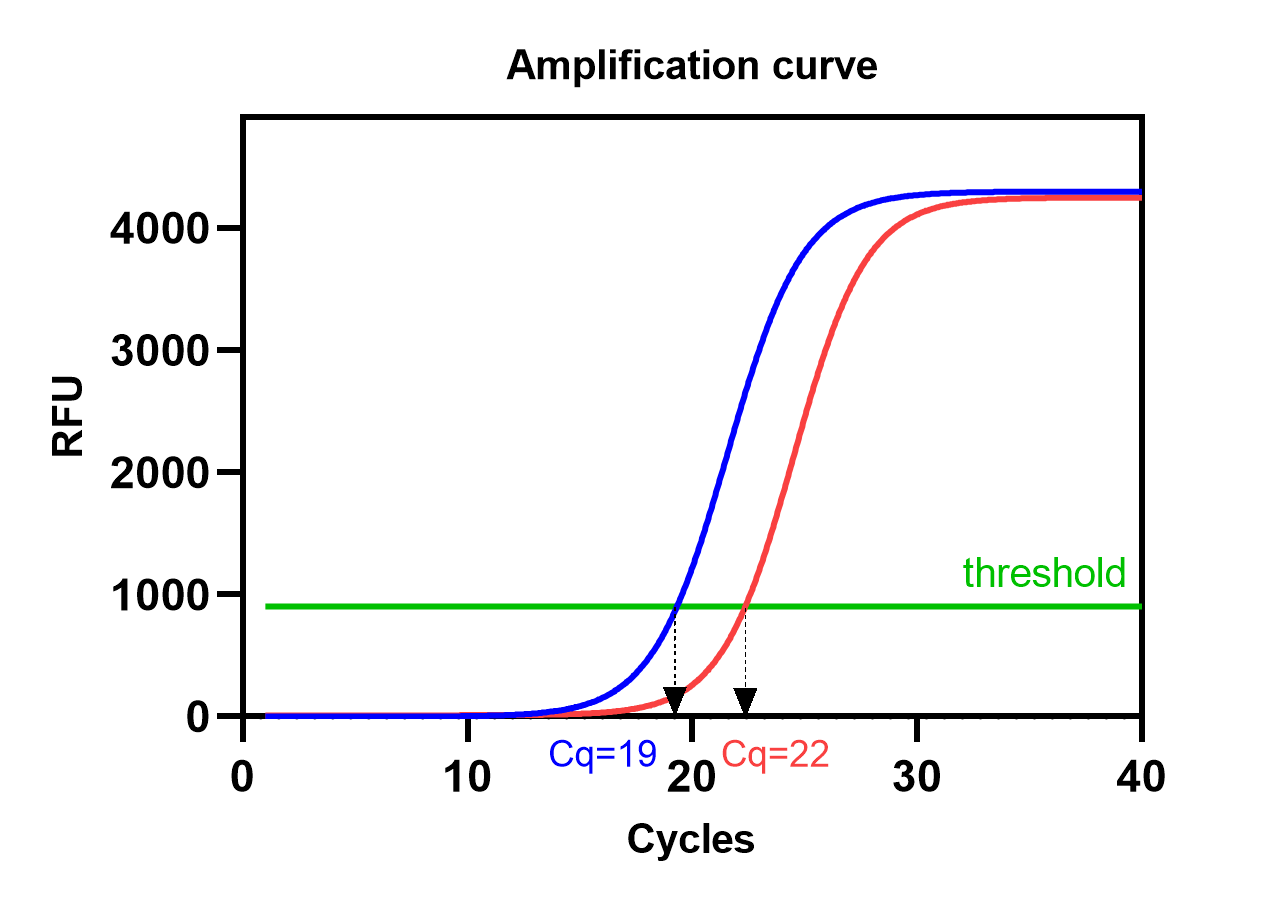
A qPCR run consists of around 40 cycles (Cq). During each cycle, three temperature stages are repeated resulting in amplification of the amount of target cDNA (figure 1). One Cq difference means that there is a twofold (Log2) difference in the amount of target cDNA. The amount of amplified cDNA is measured after each cycle using a fluorescent probe and will be plotted in the amplification curve (figure 2). The moment the amplification curve crosses the threshold is called the Cq value. The threshold is set by the machine software itself to distinguish relevant amplification from the background signal. It is usually the moment where the exponential phase of the curve starts to become linear. The lower the Cq value, the more RNA copies of the specific gene were present in the sample.